Bioinformatics Tools
MitoFinder
Allio, R., Schomaker‐Bastos, A., Romiguier, J., Prosdocimi, F., Nabholz, B., & Delsuc, F. (2020). MitoFinder: Efficient automated large‐scale extraction of mitogenomic data in target enrichment phylogenomics. Molecular Ecology Resources, 20(4), 892-905. DOI: 10.1111/1755-0998.13160
Mitofinder is a pipeline to assemble mitochondrial genomes and annotate mitochondrial genes from trimmed read sequencing data. MitoFinder is also designed to find and annotate mitochondrial sequences in existing genomic assemblies (generated from Hifi/PacBio/Nanopore/Illumina sequencing data…)
PhylteR
Comte, A., Tricou, T., Tannier, E., Joseph, J., Siberchicot, A., Penel, S., Allio, R., Delsuc, F., Dray, S., & de Vienne, D. M. (2023). PhylteR: efficient identification of outlier sequences in phylogenomic datasets. Molecular Biology and Evolution, Volume 40, Issue 11, November 2023, msad234. DOI: 10.1093/molbev/msad234
PhylteR is a tool that allows detecting, removing and visualizing outliers in phylogenomics dataset by iteratively removing taxa from gene families (gene trees) and optimizing a score of concordance between individual matrices.
Database
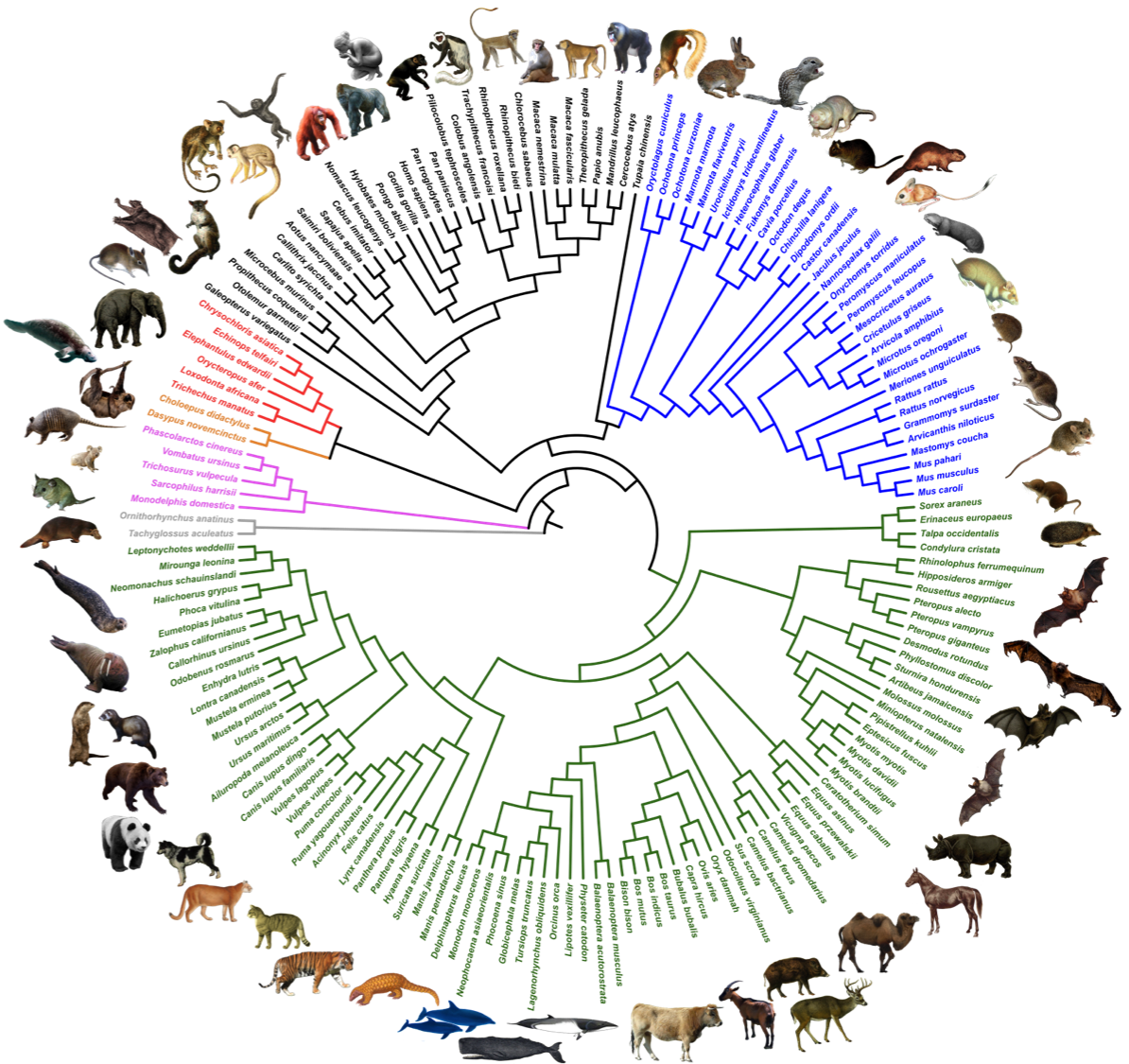
OrthoMaM
Allio, R., Delsuc, F., Belkhir, K., Douzery, E. J., Ranwez, V., & Scornavacca, C. (2023). OrthoMaM v12: a database of curated single-copy ortholog alignments and trees to study mammalian evolutionary genomics. Nucleic Acids Research, gkad834. DOI: 10.1093/nar/gkad834
OrthoMaM , a database of ORTHOlogous MAmmalian Markers describing the evolutionary dynamics of coding sequences in mammalian genomes. OrthoMaM version 12 includes 15,868 alignments of orthologous coding sequences (CDS) from the 190 complete mammalian genomes currently available.
Protocols
ONT sequencing
Allio R., Tilak M. K., Scornavacca C., Avenant N. L., Corre E., Nabholz B., & Delsuc F. (2021). High-quality carnivore genomes from roadkill samples enable species delimitation in aardwolf and bat-eared fox. eLife, 10, e63167. DOI: 10.7554/eLife.63167
This protocol was developed to optimize DNA sequencing from mammalian roadkill tissue samples using Oxford Nanopore Technology (ONT).